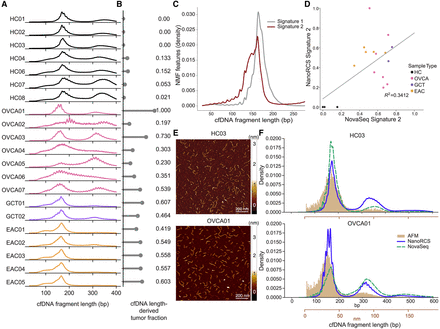
NanoRCS captures cfDNA fragmentation length distribution indicating tumor presence. (A) cfDNA fragmentation length profiles for each sample, categorized by sample type. (HC) healthy controls (black), (OVCA) ovarian carcinoma (pink), (GCT) granulosa cell tumor (purple), (EAC) esophageal adenocarcinoma (orange). The profiles display the distribution of cfDNA fragment sizes ranging from 30 to 400 bp. (B) TF for each sample derived by mapping the length profile to Signature 2 of the nonnegative matrix factorization (NMF) on a reference set (see Methods). TF is annotated to the right of each bar, ranging from 0.14 to 1.0 for tumor samples. (C) Representative NMF cfDNA length profiles adapted from Renaud et al. (2022). for two signatures: Signature 1, predominantly observed in healthy individuals, and Signature 2, indicative of tumor-derived cfDNA. (D) Scatter plot correlating the fragmentomics NMF-derived TF of NanoRCS and NovaSeq against each other (R2 = 0.3412). (E) Atomic force microscopy (AFM) images visualizing HC03 (top) and OVCA01 (bottom) cfDNA fragments (yellow). Scale bar, 200 nm. (F) Density plots comparing the length distribution obtained from three different techniques (blue, NanoRCS; green, NovaSeq; yellow, AFM) in sample HC03 (top) and OVCA01 (bottom). Conversion of the x-axis in nm (brown) to the x-axis in bp (black) is based on the calculation of the DNA ladder where Lbp = (Lnm + 10)/0.341. AFM imaging (brown) appears to better represent the shorter fragments, while both sequencing methods enriched for longer fragments, especially NanoRCS (blue).











