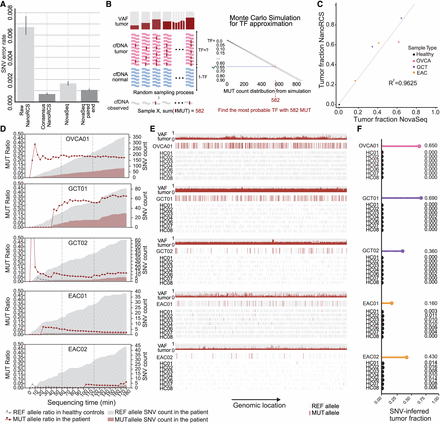
NanoRCS enables SNV detection with a low error rate on Nanopore. (A) Single-nucleotide error rate in cfDNA of three HCs using different sequencing methods (lower is better). Error bars represent standard deviations. (B) Monte Carlo simulations are used to search for the TF that best explains the observations in cfDNA given the known tumor variants and their variant allele frequency (VAF) in the tumor. Created with BioRender (for detailed methods, please see Methods: “Tumor fraction estimation from somatic SNV detection”). (C) Correlation of SNV-derived TF between NanoRCS and NovaSeq. (D) Real-time assessment of the ratio of the mutant (MUT; red shading) and reference (REF; gray shading) allele at somatic SNV positions during the first 3 h of sequencing in the five liquid biopsy samples with known somatic SNV profile from the tumor biopsy. Red data points and lines indicate the MUT SNV ratios in cancer patients, and dark gray data points and lines in HCs (at level ∼0.00). (E) SNV observations in the five liquid biopsy samples with known tumor somatic SNV profile. For each panel, the top row shows the VAF of detected mutations in the tumor biopsy, the second row represents the MUT or REF allele observations in the liquid biopsy of the corresponding patient, and the bottom three rows represent the observations in three HCs randomly downsampled to the same amount of observation as in the tumor sample. (F) Lollipop plots show each sample's inferred TF according to corresponding tumor-informed variants in C.











