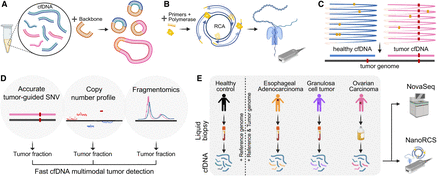
NanoRCS can identify SNVs, CNVs, and fragment size in cfDNA. (A) Healthy cfDNA (blue) and tumor ctDNA (pink) are circulated with a high curvature DNA backbone (orange) and form a double-strand circular DNA product. (B) DNA primers of random sequence and phi29 polymerase are added to these DNA circles. RCA of double-strand circular DNA templates subsequently creates long concatemers of cfDNA and backbone that are sequenced on a nanopore device. (C) Obtained DNA sequences are aligned to the reference genome and undergo consensus calling, resulting in cfDNA consensus sequence with reduced random errors (yellow dots) and retained true variants (red dots). After consensus calling, the tumor mutations can be found in the tumor cfDNA and not in the healthy cfDNA. (D) NanoRCS allows simultaneous assessment of fragmentomics, copy number profile, and accurate tumor-guided SNVs. ctDNA fraction can be derived based on all three modalities. (E) cfDNA from plasma of healthy controls, EAC and granulosa cell tumor (GCT) patients, and cfDNA from ascites of ovarian carcinoma (OVCA) patients is subjected to genome-wide NanoRCS. We also sequenced the same patient samples with Illumina NovaSeq. Created with BioRender (https://www.biorender.com).











