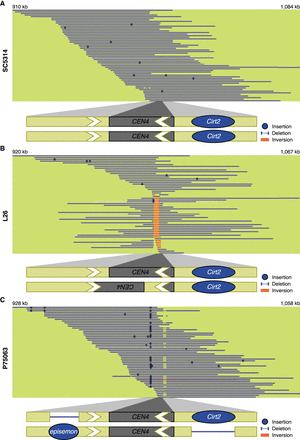
SVs in the pericentromere of Chr 4 in clinical isolates. Representative long reads aligned to CEN4 in the C. albicans reference genome for (A) SC5314, (B) L26, and (C) P75063. Binding of the centromere-specific histone H3 variant Cse4p/CENPA delineates the central core sequence of the centromere and is indicated with a gray box (Sanyal et al. 2004; Ketel et al. 2009). Blue lines indicate reads in the same orientation as the reference genome, while red reads indicate an inverted orientation compared to the reference genome. Insertions and deletions compared to the reference genome are denoted with blue dots or a thinner blue line, respectively. Insertions that are not shared by more than one reads are considered to be sequencing errors. The schematic at the bottom represents a model of the CEN4 structure (lengths not to scale). Blue circles indicate the presence of a TE. White arrows indicate inverted repeat sequences identified previously (Selmecki et al. 2006; Todd et al. 2019).











