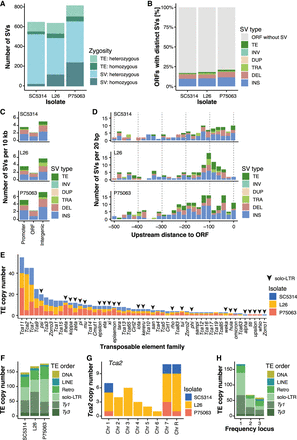
Genome-wide distribution of SVs. (A) Zygosity of SVs and TEs. The bar plot represents the copy number and zygosity for SVs and TEs in the three isolates. (B) Open reading frames (ORFs) close to SVs and TEs: Color indicates the percentage of ORFs that either overlap with at least one SV or TE, or that have at least one SV or TE in a 500 bp window upstream or downstream from the ORF. Gray indicates that no SV or TE is present. Color by SV type or TE: (TE) transposable element, (INV) inversion, (DUP) duplication, (TRA) translocation from another chromosome, (DEL) deletion, (INS) insertion. (C) SVs and TEs by distance to the ORF: location in the promoter region (500 bp upstream of the ORF), overlapping with the ORF, or intergenic. Color by SV type or TE. Corrected by the number SV per 100 kb, as the three different regions cover space of different lengths. (D) Fine-scale SV and TE distribution in the promoter region (500 bp upstream of the ORF). Color by SV type or TE. (E) Frequency of TE families between the three isolates. A triangle indicates this element is a solo-LTR. The sequences and their corresponding full-length elements can be found in Supplemental Table S1. (F) TE copy numbers in the three clinical isolates. Colors indicate the classification (Ty1 and Ty3 are superfamilies in the LTR-retrotransposons; DNA and retrotransposon are on the class level; LINE is the order of non-LTR-retrotransposons; solo-LTR are fragments of Ty1 or Ty3, and unknown retrotransposons). (G) Copy number of the Tca2 transposon on each of the C. albicans chromosomes. Colors indicate the isolate. (H) Frequency distribution of TE loci that are identical in location between the three isolates. Colors indicate the classification.











