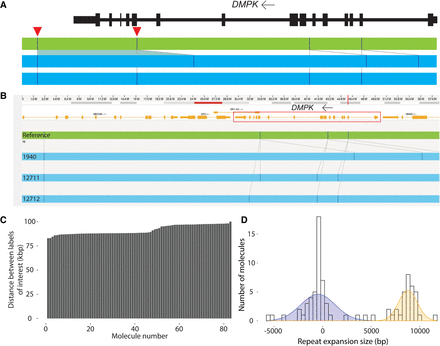
Overview of the data analysis outputs of the three OGM repeat expansion workflows for sample DMPK_10.This figure only shows the visual results of the data analysis. The results of the data interpretation are mainly the estimates of the actual repeat sizes resulting from the manual de novo assembly and local-GA workflows, as well as the visualization of the label distances in each molecule covering the locus of interest resulting from the molecule distance script. (A) Representation of the repeat expansion locus in the de novo assembly showing the position of the repeat expansion in the gene (3′ UTR). Labels of interest are indicated by red arrowheads. These labels were used to manually calculate the repeat size by subtracting the reference distance (green bar) from the distances of the respective sample maps (blue bars). (B) Consensus-guided assemblies across the DMPK repeat expansion locus. The DMPK gene is indicated by the red box. Based on the estimated repeat length, each map is assigned to allele 1 or allele 2 in order to separate the two alleles. Final repeat sizes are calculated by combining the repeat sizes of the maps assigned to the same allele (see also Methods). (C) This bar plot shows the distance between the labels of interest in each molecule ordered from smallest to largest. (D) This histogram shows the result of the molecule distance script that automatically assigns molecules to one of the alleles. The blue peak represents allele 1, while the orange peak represents allele 2. Both the bar plot and histogram can then be used to assess whether a sample contains evidence for somatic instability or not.











