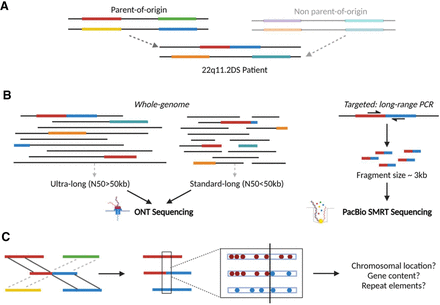
Study design and data analysis of 22q11.2DS recombination loci. (A) LCR22 composition of a family trio. The parent-of-origin is the parent in whom the recombination occurred. The red-blue composition in the patient represents the recombined LCR22 structure. In this example, an interchromosomal recombination is presented, although intrachromosomal recombinations are possible as well. (B) The patient and the parent-of-origin were sequenced using one or a combination of whole-genome ultra-long and/or standard long ONT sequencing. In one patient (AB004), a targeted breakpoint-specific long-range PCR was designed in combination with PacBio single-molecule real-time (SMRT) sequencing of the fragment. (C) Following de novo assembly of the different proximal and distal LCR22 alleles in the patient and parent-of-origin, the recombination-involved parental haplotypes (proximal and distal) were identified via SNP comparison, followed by a delineation of the breakpoint locus by LCR22 proximal-specific (red) and distal-specific (blue) SNPs. These SNPs are shared between the patient and the parental proximal or distal LCR22, respectively, but not with the other (proximal or distal) LCR22 involved in the CNV. The recombination locus was scrutinized for its precise coordinates, elucidating the genes and repetitive elements involved.











