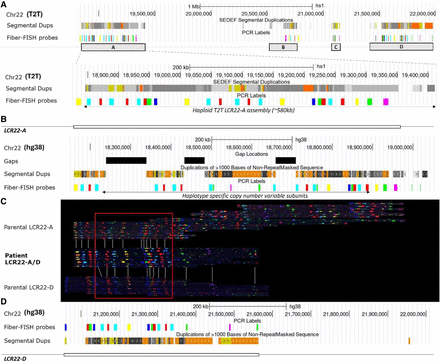
Fiber-FISH analysis of an LCR22-mediated (A–D) rearrangement. (A) UCSC Genome Browser screenshot of T2T with tracks for SDs and BLAT sequences for fiber-FISH probes in the 22q11.2 region LCR22-A to -D (up) and a close-up of LCR22-A (down). (B) UCSC Genome Browser screenshot of GRCh38 (hg38) with tracks for gaps, SDs, and BLAT sequences for fiber-FISH probes in LCR22-A. (C) LCR22-A/D rearrangement with de novo assembled parental LCR22-A, -D allele that contributed to the rearrangement, and 22q11.2DS patient rearrangement alleles. The parental LCR22-A haplotype is not identical to the reference allele in A, because of the presence of gaps in the reference and haplotype variability observed in the human population. The sequence in the red box is shared between three alleles and therefore considered to be the locus where putative recombination had taken place. Gray lines indicate similarities between the alleles of the patient and the parent-of-origin. (D) UCSC Genome Browser screenshot of hg38 with tracks for fiber-FISH probes and SDs in LCR22-D.











