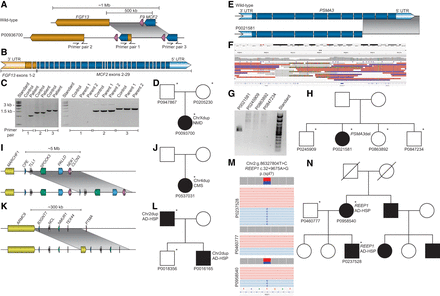
Visualization of candidate disease-causing SVs in the “unsolved” cohort. In a sporadic patient (P0093700) with arthrogryposis multiplex congenita, we detected an X-Chromosomal tandem duplication (A–D). The duplication spans from intron 1 of MCF2 to intron 2 of FGF13 and also includes F9 (A). The result of the duplication is a hypothetical fusion gene, including FGF13 exons 1–2 and MCF2 exons 2–29 (B). The duplication was validated by PCR and agarose gel electrophoresis (C) using primers targeting the breakpoints of the duplication, and a combination of the MCF2 forward and FGF13 reverse primers (primer pair 1, A,C). In a sporadic patient with psychomotor development delay (P0021581), we detected a deletion of PSMA3 exons 9–11, here shown as a cartoon (E) and as a screenshot using IGV (F). The deletion was validated by agarose gel electrophoresis (G) and Sanger sequencing. The index P0021581 has three healthy siblings who do not carry the deletion (H). In a sporadic female patient (P0537031) with congenital malformation syndrome, we detected a 5 Mb tandem duplication on Chromosome 4, visualized here as a cartoon (I). The pedigree is shown in J. In a sporadic male patient (P0016165) with autosomal dominant spastic paraplegia, with a similarly affected father, we detected a 300 kb tandem duplication on Chromosome 2, visualized here as a cartoon (K). The pedigree is shown in L. In a family with an affected mother and son (P0847234 and P0958540), we detected an intronic variant in REEP1 (M). The reads are colored by haplotag; pink and light blue represent different alleles in M. The index patient also has an affected uncle, whose sample was not sequenced (N). Sequenced individuals are marked with an asterisk (*) in the pedigrees (D,H,J,L,N). (NMD) Neuromuscular disorder, (CMS) congenital malformation syndrome, (AD-HSP) autosomal dominant hereditary spastic paraplegia.











