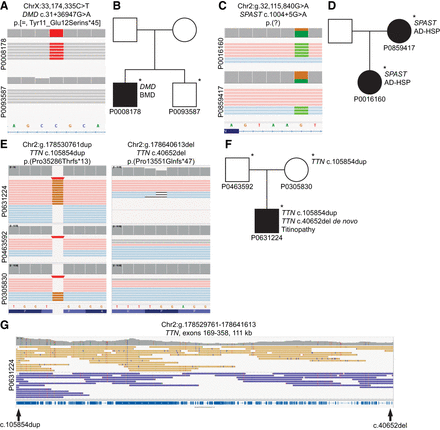
Visualization of disease-causing SNVs and indels in the “unsolved” subcohort in the form of IGV screenshots, along with corresponding pedigrees. In a sporadic patient with suspected titinopathy, we identified a deep-intronic variant in DMD (A). The nonaffected sibling did not carry the variant (A,B). In a duo consisting of an affected mother and affected daughter with HSP, we identified a noncanonical splice site variant in SPAST (C,D). In a patient with titinopathy (P063122), a maternally inherited and a de novo variant had been identified earlier (C–F). The two variants are located 109 kb apart, but the alleles were successfully phased through the entire region by LRS (G). The reads are colored by haplotag; pink and light blue, or yellow and purple represent different alleles in A, C, E, and G. Unphased reads, such as X-Chromosomal reads in males, are shown in gray (A,E). Sequenced individuals are marked with an asterisk (*) in the pedigrees (B,D,F). (BMD) Becker muscular dystrophy, (AD-HSP) autosomal dominant hereditary spastic paraplegia.











