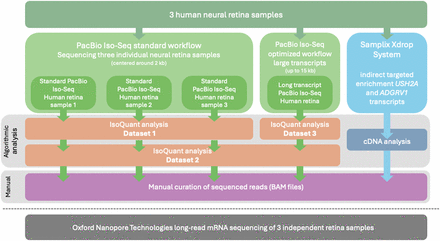
Overview of the sequencing workflows and subsequent analyses. The figure illustrates the sequencing workflows and subsequent analysis performed on RNA extracted from three human neural retina samples. The workflows include PacBio long-read mRNA Iso-Seq using both the standard and an optimized long transcript workflow. The analysis was carried out in three distinct data sets: data set 1 comprised the standard workflow samples analyzed with IsoQuant, data set 2 involved a combined analysis of the reads obtained with standard and optimized long transcript workflows, and data set 3 focused solely on reads obtained with from the long transcript workflow. Additionally, an “indirect targeted enrichment” of transcripts for the USH2A and ADGRV1 genes was achieved using the Samplix Xdrop System, followed by PacBio long-read sequencing and cDNA analysis. All reads mapping to Usher syndrome–associated transcript isoforms were manually curated using BAM files in the IGV. An independent ONT long-read sequencing data set of three independent retina samples was used to validate findings.











