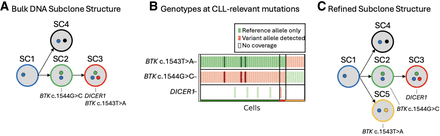
Refining the subclone structure of patient 3. (A) The subclone structure identified in the bulk DNA sequencing data of patient 3. CLL-relevant gene mutations are annotated under the subclone they are found in B. The genotype matrix plot from the relapse sample of patient 3 enables refinement of the original subclone structure. Each cell that was successfully assigned to a subclone is shown (x-axis), along with every CLL-relevant variant that was present in the WES data (y-axis). Green markers indicate that only reference alleles were present in the scRNA-seq reads at the given variant location within the cell, and red markers indicate that at least one scRNA-seq read in the cell contains the somatic variant allele. Darker coloring indicates an increased number of reads supporting that genotype. Only the CLL-relevant mutations are included for increased resolution to differentiate subclones. (C) The refined subclone structure that depicts the subclone containing the BTK c.1543T > A mutation is independent of the subclone containing the BTK c.1544G > C and DICER1 mutations.











