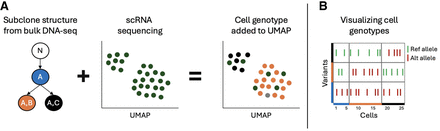
Overview of workflow to identify and use cell genotypes. (A) Predetermined subclone structures with accompanying somatic variant information are used to genotype individual cells in scRNA-seq data. Cells are assigned to a predetermined subclone based on the presence or absence of subclone-defining mutations. Subclone assignments are then used to group cells of the same subclone to identify subclone-specific gene expression. (B) Genotype matrix plots visualize the genotypes of all cells at each variant of interest, showing green for reference allele, red for alternate allele, and white for no coverage.











