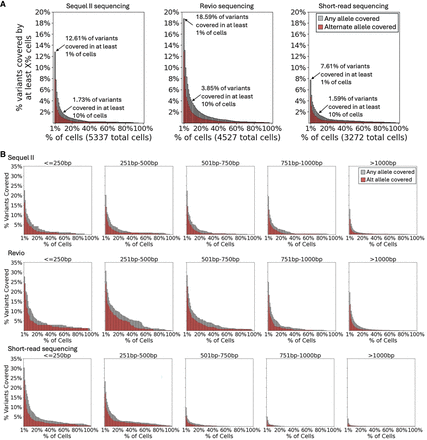
The variant coverage provided by each scRNA-seq technology. (A) The overall variant coverage provided by the Sequel II and Revio compared to Illumina short reads. The percentage of cells covering each heterozygous germline variant in the patient's WES data is used to determine the percent of variants covered by at least X% of cells. (B) The variant coverage is binned by the variant's distance from the priming site, as indicated above each plot (bp = base pairs).











