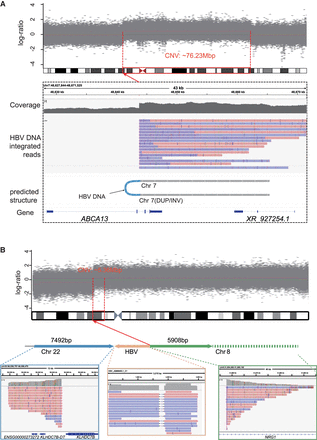
Inferring SV types based on HBV DNA integration patterns. (A) In sample HCC9_T3, an HBV DNA INS induces a fold-back INV in the host DNA. The top panel illustrates the CNV increase occurring ∼76.23 Mb downstream from the integration site. Sequencing data demonstrate that long reads encompass the sequences flanking the HBV integration, revealing identical breakpoints and reverse DUP of the adjacent sequences. (B) An example of HBV DNA integration resulting in a TRA (HCC13_T2). The Integrative Genomics Viewer (IGV; Robinson et al. 2011) plot below illustrates the alignment of long reads to Chr 22, HBV, and Chr 8. The plot above depicts the concordance of its breakpoint on Chr 8 (red dashed line) with a breakpoint of ∼5.36 Mbp CNV (gray shade).











