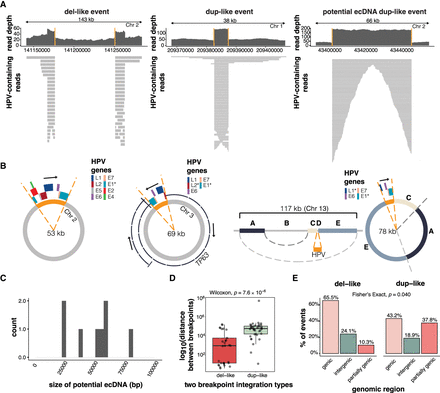
The characteristics of two-breakpoint events and potential ecDNAs. (A) Examples of read coverage patterns from a del-like event, a dup-like event, and a potential ecDNA dup-like event. Orange lines indicate the integration breakpoints. (B) Circular assemblies from three potential ecDNA integration events. The orange portions show the integrated HPV segment, including the viral genes. The direction of HPV gene transcription is shown by a black arrow. The right-most example depicts a complex event in which three nonadjacent human segments have been combined in the potential ecDNA. (C) The size distribution of potential HPV–human hybrid ecDNAs (n = 8). (D) The genomic distance between breakpoints in del-like versus dup-like events. Box plots represent the median and upper and lower quartiles of the distribution; whiskers represent the limits of the distribution (1.5 IQR below Q1 or 1.5 IQR above Q3). (E) The percentage of events occurring in genic (>90% within a gene), intergenic (<10% within a gene), and partially genic (10%–90% within a gene) regions, plotted by integration category. The P-value in D was calculated using a Wilcoxon ranked-sum test. The P-value in E was calculated using a Fisher's exact test.











