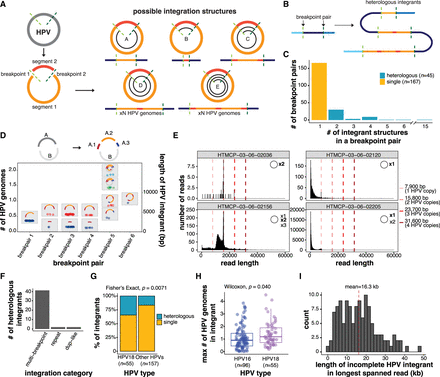
Heterologous structures of the HPV genome before and after integration. (A) Schematic of possible HPV integrant structures. The spirals in structures A–E show the portion(s) of the HPV genome that could be contained within an integrant. (B) Schematic showing how several heterologous integrants can exist between a single breakpoint pair, with the size of the integrant varying by n HPV copies. The colors correspond to the regions of the HPV genome as depicted in A. (C) The number of integrant structures between all identified breakpoint pairs within the cohort. Integrants with 2+ identified structures were classified as heterologous. (D) The sizes of the HPV integrants in a multi-breakpoint event with schematics depicting the various integrant structures detected. The HPV genome, in this case, was broken into two segments A and B, and the A segment was further broken into three segments (A.1, A.2, and A.3). These segments were variably rearranged into new structures across the breakpoint pairs. Each point represents the size of an HPV integrant contained on an individual read, which is then grouped together by color (e.g., blue, red, green, light blue) if they do not differ in size by more than 300 bp. Each color in a breakpoint pair thus represents one unique integrant structure, as indicated in the accompanying schematics. (E) The lengths of HPV-aligned reads in four predominantly episomal samples. The existence of HPV episomes and episome concatemers is supported by the accumulation of read counts in bin sizes corresponding to one or more HPV genome copies, as indicated by the dotted lines. (F) Frequency of heterologous integrants in the different integration categories. Only categories harboring heterologous integrants are shown. (G) The percentage of integrants from different HPV types that form single or heterologous structures. (H) The maximum size of the integrant structure in each breakpoint pair in HPV16 and HPV18 integrants, represented as the number of HPV genome copies. (I) Distribution showing the maximum number of HPV copies found in the longest spanning read for each incomplete integrant. The x-axis shows the length of the longest spanning read. Box plots represent the median and upper and lower quartiles of the distribution; whiskers represent the limits of the distribution (1.5 IQR below Q1 or 1.5 IQR above Q3). P-values were calculated using the Wilcoxon rank-sum test and the Fisher's exact test, as indicated in the figure.











