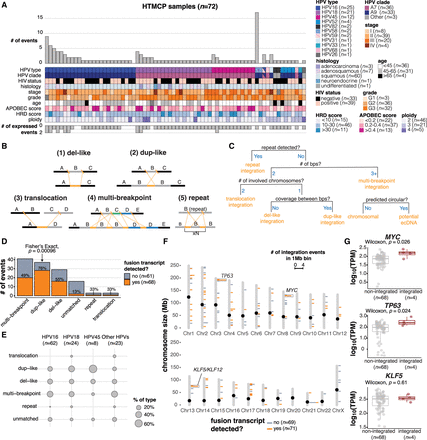
Detection and categorization of HPV integration events in cervical cancers. (A) The number of HPV integration events detected in the DNA (# of events) and detected in DNA and RNA (# of expressed events) across the HTMCP samples, as well as clinical and molecular characteristics. (B) Schematic illustrations of integration event categories. Orange segments indicate the insertion of HPV integrants. (C) Decision chart for categorizing HPV integration events (“bps” = breakpoints). (D) The frequency of the HPV integration categories across the samples. The percentage of events that produce an HPV–human fusion transcript is indicated for each integration type. (E) The percentage of events belonging to each integration category for HPV16, HPV18, HPV45, and all other HPV types. (F) The genomic locations of integration events across the cohort, colored by the transcriptional status of the event. Bins with 2+ integration events are highlighted with boxes. Notable cancer genes within bins recurrently affected by integration are indicated. (G) Gene expression differences between samples with HPV integration within 1 Mb of MYC, TP63, and KLF5 compared to the remaining samples in the data set. Box plots represent the median and upper and lower quartiles of the distribution; whiskers represent the limits of the distribution (1.5 IQR below Q1 or 1.5 IQR above Q3). P-values were calculated using the Wilcoxon rank-sum test.











