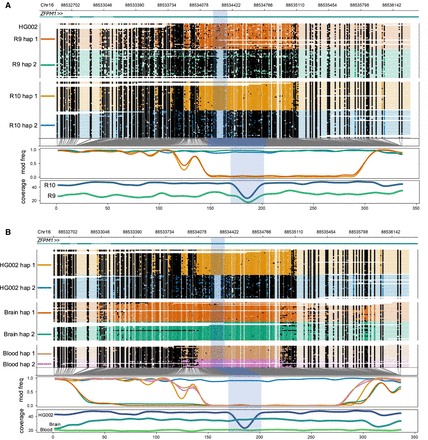
Haplotype-specific methylation differences and similarities between cell, blood, and brain samples. From top to bottom, each plot shows the genome coordinates, labeled gene models (if present), haplotype-aware read mappings with modified bases as black (methylated), or colored (unmethylated) circles, a smoothed methylation fraction plot, and a coverage plot. The highlighted region corresponds to a 75 bp deletion (Chr16:88,534,247–88,534,321) in haplotype 2 of the HG002 cell line that coincides with haplotype-specific methylation. Coordinates across the bottom refer to methylation bins used in the smoothed methylation plot. (A) Haplotype-specific methylation differences and similarities between the R9 and R10 sequenced HG002 cell line. (B) Haplotype-specific methylation differences and similarities between the R10 sequenced cell line, blood, and brain samples.











