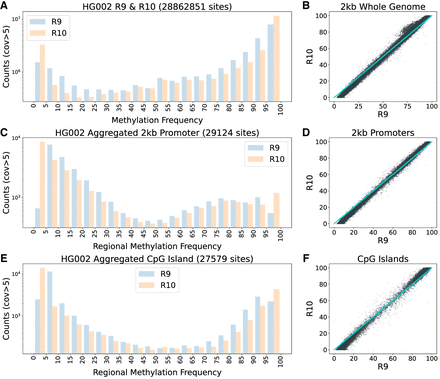
Genome-wide and regional methylation calling trends in HG002 R9 and R10 data sets. (A) Methylation calls for all CpG motifs (strand combined) in HG002 R9 and R10. (B) Scatterplot of 2 kb whole-genome windows with a minimum of 10 CpGs for R9 and R10. The 0–100 diagonal line is overlaid in cyan for reference. (C,D) Average methylation over the 29,124 filtered human promoters with a minimum of 10 CpGs plotted as a histogram and a scatter plot. (E,F) A histogram and scatter plot of the CpG island BED regions with more than 10 CpGs in HG002. All plots are limited to CpG sites with >5× read coverage. The y-axis is log scaled.











