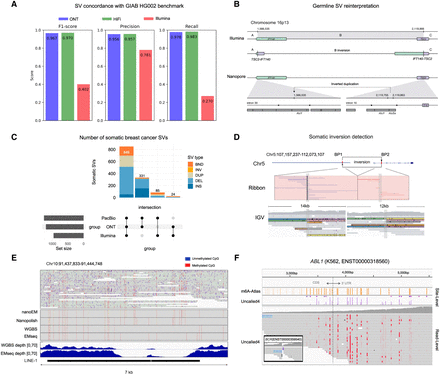
Comparative analysis of structural variation and epigenetics using short- and long-read sequencing. (A) Performance comparison of structural variant detection across sequencing technologies, evaluated against the GIAB Tier1 V0.6 benchmark (Kolmogorov et al. 2023). (B) Reinterpretation of a germline recurrent event using long-read sequencing. (Upper panel) Illumina data indicate a long-range inversion on Chromosome 16p13 with breakpoints in IFT140 and TSC2. (Lower panel) ONT sequencing reveals the actual structure: an insertion in intron 30 of IFT140 (Thibodeau et al. 2020). (C) UpSet plot of somatic structural variants identified in the breast cancer cell line using three sequencing technologies (Keskus et al. 2024). (DEL) Deletion, (BND) breakend junction, (DUP) duplication, (INV) inversion, (INS) insertion. (D) 4.9 Mbp somatic inversion detected by nanopore sequencing in colorectal cancer, encompassing exon 1 of the APC gene (Xu et al. 2023). (E) Methylation patterns of LINE-1 located in HECTD2 intron. (Upper panel) Reads from breast cancer cell lines aligned to the region. (Lower panel) Read coverage of EMseq and WGBS for the same region (Sakamoto et al. 2021a). (F) m6A profiling of an ABL1 transcript and BCR-ABL1 fusion in chronic myeloid leukemia (CML) (Kovaka et al. 2024). (C–F, adapted from Thibodeau et al. 2020, Sakamoto et al. 2021a, Xu et al. 2023, and Kovaka et al. 2024).











