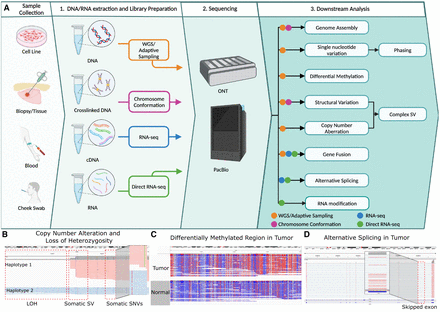
Overview of long-read sequencing protocols and analyses. (A) The overview of the long-read sequencing workflow including downstream analysis options. (B) Somatic SNV and a structural variation in the COLO829 cell line resulting in haplotype-specific copy number change and loss of heterozygosity (data from Keskus et al. 2024). Reads are grouped and colored with the alleles using long-read-based phasing. (C) Differentially methylated region between tumor and normal sample. Reads are colored with 5mC modifications (red: high methylation, blue: low methylation). (D) An alternative splicing in CASC4 gene in cancer cell line SCC152 (Rodriguez et al. 2024).











