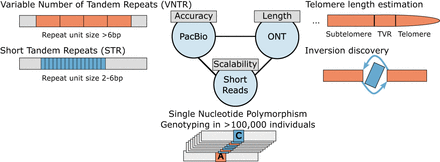
Long-read sequencing facilitates accurate discovery of SVs and their breakpoints at single-nucleotide resolution, with PacBio HiFi sequencing showing very high accuracy in terms of mapping repeat polymorphisms and ONT ultra-long (ONT-UL) being an essential method for determining centromeric or telomeric variant repeat (TVR) structures as well as balanced inversions which are often embedded within large DNA repeats. Short reads remain highly cost-efficient and thus, scalable to tens of thousands of genomes. Notably, some applications such as telomere-to-telomere assembly projects presently require a combination of technologies, with current studies using PacBio Hifi sequencing, ONT-UL, and Strand-seq (short reads) (Logsdon et al. 2024a).











