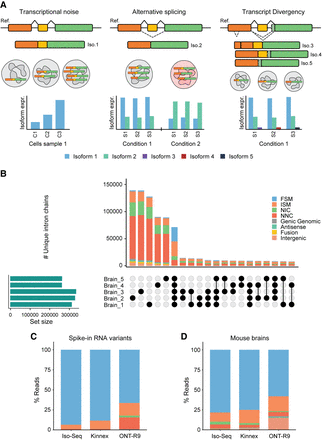
TD detection. (A) Differences between TN, alternative splicing (AS), and TD. TN indicates stochastic variation in transcript expression levels. AS is programmed alternative processing of RNA that can be consistently detected. TD represents rare but real RNA molecules. (B) Structural category classification and intersection of loci among biological replicates in mouse brain samples. Replication strongly reduces the biological and technical noise in the samples. Structural category classification of reads among PacBio Iso-Seq and Kinnex, and ONT (R9 flowcell) sequencing methods in (C) spike-in RNA variants (SIRVs) and (D) mouse brains. Non-FSM reads in spikes represent the technical noise level of the technology, whereas the excess of these reads in the real sample is indicative of TD.











