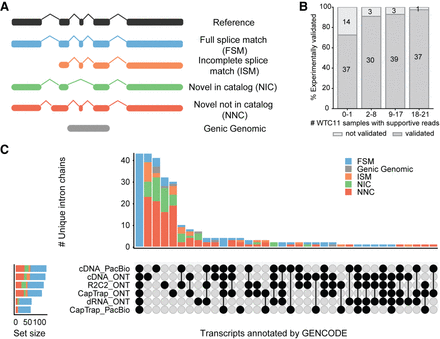
LRGASP identifies many transcripts only expressed in one or a few samples. (A) Main SQANTI3 structural categories for transcript models of known genes. (B) Fraction of experimentally validated transcripts as a function of the number of WTC11 samples in which supportive reads were observed. (C) Structural category classification and intersection of 50 loci from the WTC11 samples that were not identified by any transcript identification tool but were manually annotated by GENCODE. These loci were selected for having mapped reads across all six library preparation and sequencing platform combinations. (B,C) Adapted from Pardo-Palacios et al. (2024b).











