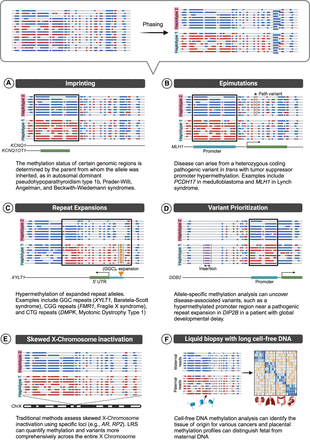
Diagnostic applications of haplotype-aware methylation analysis. Haplotype assignment after phasing significantly enhances the diagnostic utility of LRS. By enabling the simultaneous detection of SNVs, SVs, CNVs, and DNA modifications, it allows disease mechanisms that previously required separate specialized tests to be studied under a single, unified approach, such as imprinting (A), epimutations (B), and repeat expansions (C). It also facilitates variant prioritization (D), enables the investigation of skewed XCI where, instead of mixed inactivation, one haplotype is inactivated consistently (E), and can even be applied to study cell-free DNA (F). Created in BioRender. Montano, C. (2024). https://BioRender.com/h78t893.











