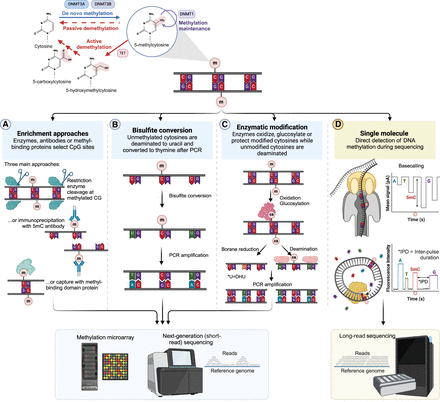
Methods to detect genome-wide 5mC. DNAm can be detected through enrichment, bisulfite conversion, enzymatic modification, or single-molecule approaches. (A) Enrichment methods use restriction enzymes or immunoprecipitation to separate methylated and unmethylated DNA. (B) Bisulfite treatment converts unmethylated cytosine to uracil, allowing differentiation between methylated and unmethylated DNA that can be assayed using microarrays or short-read sequencing. (C) Newer enzymatic methods use a series of proteins that oxidize and glycosylate modified cytosines, followed by deamination that is achieved either through specific enzymes or borane reduction. (D) Single-molecule methods directly measure DNAm as DNA passes through the nanopore (Oxford Nanopore, top panel) or as fluorescent nucleotides are incorporated into the growing strand by a polymerase (PacBio, middle panel). Created in BioRender. Montano, C. (2024). https://BioRender.com/i42h077.











