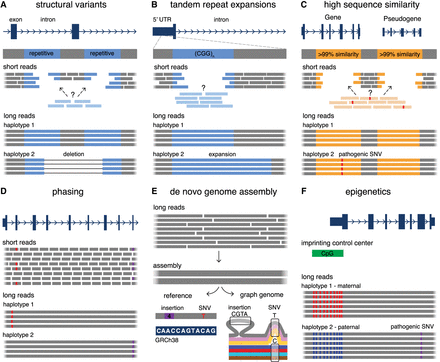
Summary of the utility of LRS over SRS in undiagnosed RDs. (A) LRS has an improved ability to detect SVs compared to SRS, especially in challenging regions for SRS such as repetitive DNA (blue), which often mediates the formation of SVs. (B) LRS enables improved sequencing and alignment of short tandem repeat sequences (blue) compared to SRS, enabling the accurate detection of tandem repeat expansions. Examples of genes with disease-associated short tandem repeat expansions include FMR1 (Fragile X syndrome), HTT (Huntington disease), and several genes associated with cerebellar ataxias (ATXN3, FGF14, etc.). (C) LRS allows for improved mapping and coverage of regions of high sequence similarity in the genome (orange), which are challenging for SRS. This enables the differentiation of sequences between genes and their pseudogenes, and therefore detection of variation in these challenging genes. Examples of disease-associated genes in regions of high sequence similarity include PKD1 (polycystic kidney disease), IKBKG (X-linked immunodeficiencies), and SMN1 (spinal muscular atrophy). (D) LRS enables haplotype phasing over long ranges, which is helpful to confirm compound heterozygosity of variants (red, purple) without the requirement of parental samples for segregation, or in scenarios where one or more variants are de novo. (E) Long reads derived from LRS can be used to build high quality and highly contiguous de novo genome assemblies without requiring alignment to a reference genome. These assemblies can either be compared to a linear reference genome (bottom left) to detect variants (a 4 bp insertion compared to the reference, purple box; and a C > T SNV, red) or they can be compared to de novo assemblies derived from other individuals (bottom right, colored lines indicate assemblies from different individuals) for the generation of graph-genomes that describe genetic variation among individuals (4 bp insertion seen only in one individual; T/C SNV that is more common in the group). (F) Sequencing of native DNA strands in LRS enables concomitant assessment of base modifications, such as differentiating between methylated cytosines (red) and unmethylated cytosines (blue) in cytosine-guanine dinucleotides (CpGs). This can be used in combination with phasing information to investigate imprinted loci that have parent-of-origin-specific DNA methylation patterns. Examples of disease-associated imprinted genes include H19/IGF2 (Silver–Russell syndrome), UBE3A (Angelman syndrome), and PLAGL1 (transient neonatal diabetes mellitus).











