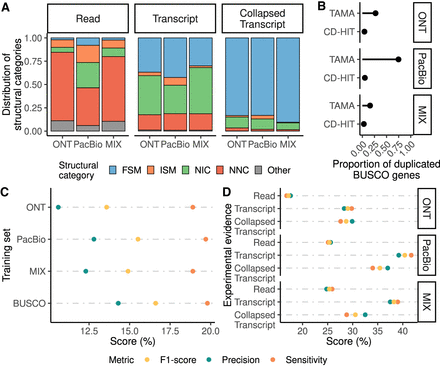
Assessment of the incorporation of long-read data to evidence-driven annotation. (A) Distribution of SQANTI3 categories at the read unique splice-junctions combination, transcript, and collapsed transcript level for PacBio data processed using Iso-Seq3 and ONT and ONT + PacBio data processed using FLAIR. (FSM) Full-Splice-Match, (ISM) Incomplete-Splice-Match, (NIC) Novel-in-Catalog, (NNC) Novel-Not-in-Catalog. (B) Redundancy of the generated transcriptome and the final collapsed transcripts set based on the proportion of duplicated BUSCO genes. (C) Evaluation of gene predictions by the three models trained with long-read data and the model trained using BUSCO genes. (D) Selection of the different long-read-based extrinsic evidence sources used for the evidence-driven gene predictions. Reads of PacBio, ONT, and a MIX of both (Read); transcript models of PacBio, ONT, and a MIX of both (Transcript) and collapsed transcripts identified in those transcriptomes (Collapsed Transcript) were used.











