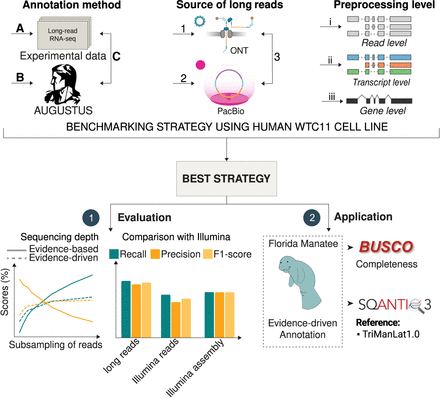
Overview of the evidence-driven, ab initio, and evidence-based annotation using long-read technologies. We compared three genome annotation strategies, evidence-based annotation using lrRNA-seq data (A), ab initio gene prediction using AUGUSTUS (B), and evidence-driven annotation combining both (C). We tested Nanopore (1) and PacBio (2) data independently and in combination (3) with processing levels ranging from raw reads (i) to transcripts (ii), and collapsing to the gene level (iii). For the best strategy defined in our benchmark, we evaluated the effect of the sequencing depth and compared to evidence-driven annotation using Illumina short-read data. Finally, we applied our strategy to a nonmodel species, the Florida manatee, applying BUSCO and SQANTI3 to evaluate the completeness of our annotation and comparing to the available Florida manatee annotation.











