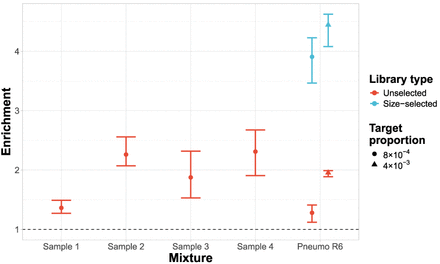
Enrichment of 23F CBL across samples containing mixed cultures from nasopharyngeal swabs. All nasopharyngeal samples (denoted “Sample X”) were run without size selection, with control samples containing Spn23F mixed with S. pneumoniae R6 (denoted “Pneumo R6”) without size selection at 0.1 and 0.5 proportions (8 × 10−4 and 4 × 10−3 23F CBL DNA proportions, respectively) run alongside. Equivalent control samples from a run with size selection are plotted for comparison. Bar ranges are interquartile range of enrichment from 100 bootstrap samples of reads. Data points are observed enrichment values for each library. The dashed line describes enrichment = 1 (i.e., no enrichment has occurred).











