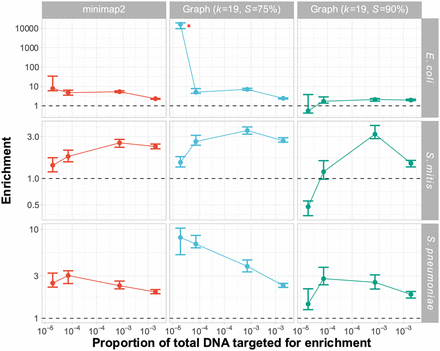
Enrichment comparison of 23F CBL at different concentrations of target between minimap2 and graph pseudoalignment in GNASTy when aligning to a partial CBL reference database. Bar ranges are interquartile range of enrichment from 100 bootstrap samples of reads. Data points connected by lines are observed enrichment values for each library, with solid lines connecting the same genome diluted at different concentrations. Rows describe the nontarget species in the mixture; columns describe the alignment method used. Each column represents data from a single flow cell. To plot on a log scale, all enrichment values had 0.01 added to them. Horizontal dashed line describes enrichment = 1 (i.e., no enrichment has occurred). Red asterisk marks high enrichment observed using graph pseudoalignment (k = 19, S = 75%) in the E. coli mixture at 4 × 10−5 target proportion.











