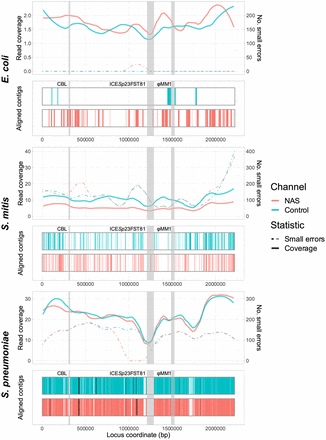
Spn23F whole-genome enrichment assembly comparison. Each panel describes an Spn23F assembly generated from 0.1 Spn23F dilutions with each nontarget organism. For each panel, the top plot shows the read coverage (solid), defined as the absolute number of bases aligning to a locus, and number of small errors (≤50 bp, dashed), and the bottom plot shows aligned contigs (colors) and large errors (black bars, >50 bp) in each assembly. Loci of interest are annotated by gray bars; CBL, as well as ICESp23FST81 and ψMM1 prophage, which are missing in this isolate of Spn23F (Croucher et al. 2012).











