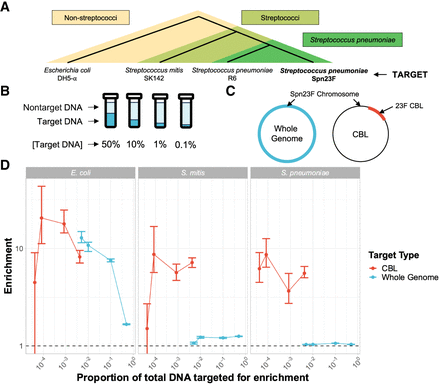
Enrichment of S. pneumoniae Spn23F in samples containing closely and distantly related nontarget species. (A) Representation of evolutionary relatedness of nontarget species and the target, S. pneumoniae Spn23F. (B) Experimental setup of target DNA dilution series with nontarget DNA. (C) Representation of two enrichment experiments, either targeting the whole Spn23F genome (blue) or the 23F CBL sequence present on the Spn23F Chromosome. (D) Enrichment results of Spn23F whole genome or 23F CBL at different concentrations of target DNA. Bar ranges are an interquartile range of enrichment from 100 bootstrap samples of reads. Data points connected by lines are observed enrichment values for each library, with solid lines connecting target DNA diluted at different concentrations with nontarget DNA. Columns describe the nontarget species within each mixture. To plot on a log scale, all enrichment values had 0.01 added to them. Horizontal dashed line describes enrichment = 1 (i.e., no enrichment has occurred).











