Table 1.
Categories of overlapping codon pairs that are the basis for codon blocks (c-blocks)
| Codon pair status | General category | C-block category | Codon topology | Description | Positional identity | Schema |
|---|---|---|---|---|---|---|
| Paired | Simple | Match | Both: contiguous or split | Exact match in genomic position and nucleotide triplet identity | 3/3 overlap |  |
| Unpaired: empty “placeholder” codon | Deletion | Both contiguous | Codon missing in the alternative isoform | 0/3 or 1/3 overlap | 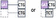 |
|
| Insertion | Codon missing in the reference isoform | 0/3 or 1/3 overlap | 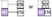 |
|||
| Unpaired: “placeholder” codon that represents an untranslated position | Translational status | Untranslated | Both: contiguous or split | Nucleotide triplet not translated in the alternative isoform | 0/3 or 1/3 overlap |  |
| Translated | Nucleotide triplet not translated in the reference isoform | 0/3 or 1/3 overlap |  |
|||
| Paired | Frameshift | Ahead | Both contiguous | Alternative isoform codon one position downstream | 2/3 overlap |  |
| Behind | Alternative isoform codon one position upstream | 2/3 overlap |  |
|||
| Split codon patterns | Edgea | Both split | Codons overlapping at the junction | Different | 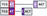 |
|
| Complexa | Both split or 1 of the 2 codons split | Codons overlapping at the junction, and are frameshifted | Different |  |
-
aA subset of these paired codons can result in AA differences, or “ragged” codon pairs.











