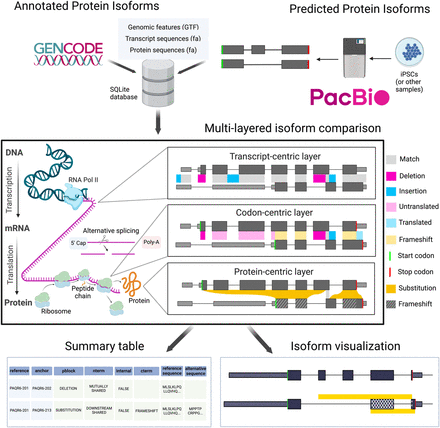
Biosurfer for analysis and visualization of protein isoform sequence differences. Biosurfer analyzes protein isoforms from reference annotations (e.g., GENCODE) or proteins predicted from long-read RNA-seq data. Analysis initiates with the creation of an SQLite database populated with isoform-relevant elements. Biosurfer performs a multilayered comparison of transcript-, codon-, and protein-level differences between pairs of protein isoforms. Variable regions, as well as their associated annotations, are output in tabular format and visualization files, which include protein-relevant details such as the frame of translation. Note that the terms “match,” “deletion,” etc., represent very different comparisons depending on the biological layer. (GTF) Gene transfer format; (iPSCs) induced pluripotent stem cells.











