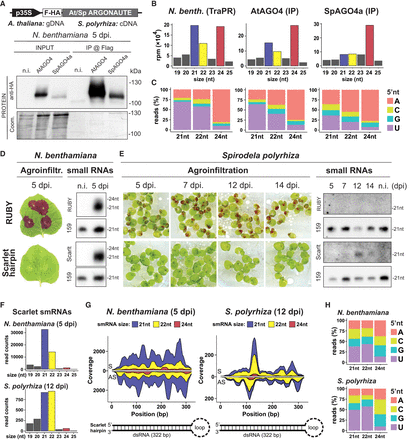
S. polyrhiza AGO4a siRNA loading preferences and IR-derived siRNAs in transient expression. (A) Protein blot analysis of Arabidopsis and Spirodela AGO4 (AtAGO4 and SpAGO4a) input and IP fractions from transient expression in N. benthamiana 5 days postinfiltration (dpi). (n.i.) Noninfiltrated. Coomassie (Coom.) staining of blot is shown as control for input and IP fractions. Scheme of constructs infiltrated in N. benthamiana leaves for transient expression is shown above. (p35S) CaMV 35S promoter, (F-HA) in-frame N-term Flag and HA peptide tags. Genomic DNA (gDNA) or cDNA origin of the AGO4 sequences is indicated. (B) Size distribution and abundance in reads per million (rpm) of TraPR purified N. benthamiana siRNAs (as control) and siRNAs extracted from AtAGO4 and spAGO4a IPs. (C) 5′-Nucleotide (5′nt) composition distribution (as percentage of reads) of 21, 22, and 24 nt siRNAs from B. (D) Representative pictures of N. benthamiana leaves agroinfiltrated for transient expression of RUBY or the scarlet hairpin (hpScarlet) IR next to sRNA blot analysis in noninfiltrated and 5 dpi leaves. miRNA159 is shown as loading control. (E) Representative pictures of Spirodela fronds agroinfiltrated for transient expression of RUBY or hpScarlet and sRNA blot analysis in noninfiltrated and infiltrated samples at different time points. miRNA159 as loading control. (F) Size distribution and abundance of sRNAs mapping to hpScarlet from 5 dpi N. benthamiana and 12 dpi Spirodela. (G) Stacked distribution of sense (S) and antisense (AS) sRNA coverage by size along hpScarlet hairpin in N. benthamiana and Spirodela. Schematic representation of the IR is depicted below. (H) 5′-nucleotide (5′nt) composition distribution of 21, 22, and 24 nt siRNAs mapping to hpScarlet in N. benthamiana and Spirodela.











