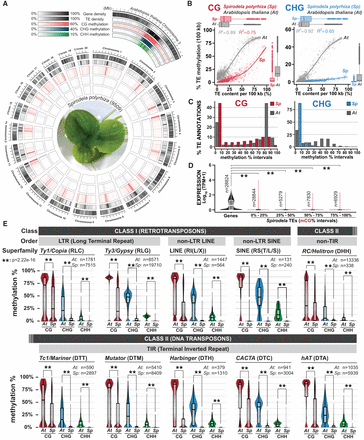
Distribution of DNA methylation in Spirodela chromosomes and TEs. (A) Circos plot (Krzywinski et al. 2009) of genome-wide distribution of DNA methylation densities by chromosome in Spirodela. From outside to inside: gene density; TE density; and CG, CHG, and CHH methylation. Mean DNA methylation, gene, and TE density values within the indicated ranges are displayed for 100 kbp intervals in 50 kbp sliding windows. Arabidopsis Chromosome 3 is shown for comparison. (B) Scatter plot and best fit line of average TE CG and CHG DNA methylation against TE content per 100 kb genomic bins in Arabidopsis and Spirodela. Colored shadows indicate the SEM. (C) Bar plot of the distribution (%) of TE annotations (≥100 bp) within 10% incremental intervals of CG and CHG methylation in Arabidopsis and Spirodela. (D) Analysis of Spirodela TE expression within 25% incremental intervals of CG methylation. Global gene expression values are shown for comparison. (E) Distribution of CG, CHG, and CHH methylation across the main TE superfamilies in Arabidopsis and Spirodela. The number of TE annotations belonging to each superfamily by species is indicated (n). (**) P-value < 2.22 × 10−16 (Wilcoxon rank-sum test). In box plots, median is indicated by solid bar; the boxes extend from the first to the third quartile, and whiskers stretch to the furthest value within 1.5 times the interquartile range. (At) Arabidopsis, (Sp) Spirodela.











