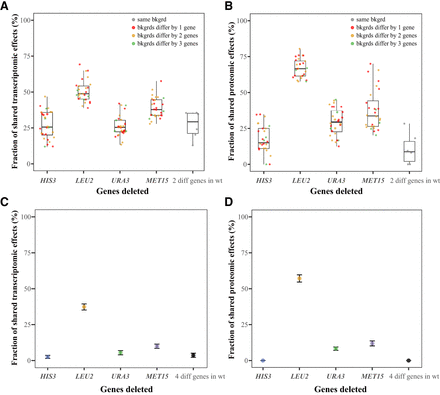
Transcriptomic and proteomic effects of gene deletions in budding yeast are sensitive to the genetic background. (A,B) Fractions of shared transcriptomic (A) or proteomic (B) effects of deleting a gene in two genetic backgrounds or deleting two different genes in the same background. Specifically, each of four focal genes (HIS3, LEU2, URA3, and MET15) is deleted in eight genetic backgrounds that vary in the presence/absence of the other three genes. For example, the focal gene HIS3 is deleted in the eight strains lacking zero to three of LEU2, URA3, and MET15. Shown for comparison are fractions of shared transcriptomic/proteomic effects of deleting two different genes in the wild-type background. In the first four bars, each dot represents a comparison between two genetic backgrounds, in which different colors indicate that the two genetic backgrounds differ by the presence/absence of one (red), two (yellow), or three (green) genes. In the last bar, each dot represents a comparison between two gene deletions in the wild type. (C,D) Fractions of shared transcriptomic (C) or proteomic (D) effects of deleting a gene between four genetic backgrounds (the wild type plus three genetic backgrounds, each lacking one of the other three genes). Shown for comparison are fractions of shared transcriptomic/proteomic effects of deleting the four different genes in the wild type. Error bars show binomial sampling-based 95% confidence intervals. In all panels, the lower and upper edges of a box represent the first (qu1) and third (qu3) quartiles, respectively; the horizontal line inside the box indicates the median (md); and the whiskers extend to the most extreme values inside inner fences; md ± 1.5(qu3−qu1).











