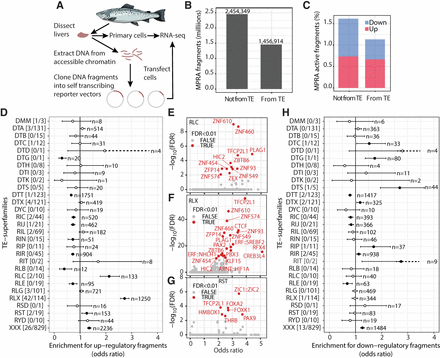
Massive parallel reporter assay screening of regulatory activity. (A) Schematic overview of the ATAC-STARR-seq MPRA experiment. (B) Barplot of the origin of sequence fragments included in the analyses. (C) Regulatory activity (inducer or repressor) of MPRA sequence fragments from TE and non-TE sequences. (D) Fisher's exact test results for enrichment of transcriptional-inducing MPRA fragments within a TE superfamily compared with all other TEs. Unknown taxonomy and DNA/retrotransposons of unknown origin (DTX/RLX) are considered separate groups. A similar test is also done on the subfamily level, and the number of significant TE subfamilies and total number of subfamilies tested are given in square brackets next to the superfamily codes. Number of regulatory active fragments are given for each category (n). (E–G) TF motif enrichment in transcriptionally inducing MPRA fragments from TE superfamilies enriched in regulatory active fragments. TF names are from the JASPAR database, and the nomenclature reflects whether it came from human or mouse. (H) Fisher's exact test results for enrichment of transcriptional-repressing MPRA fragments within a TE superfamily compared with all other TEs. A similar test is also done on the subfamily level, and the ratio of number of significant TE subfamilies to total number of subfamilies tested is given in square brackets next to the superfamily codes. Unknown taxonomy and DNA/retrotransposons of unknown origin (DTX/RLX) are considered separate groups. Number of regulatory active fragments are given for each category (n).











