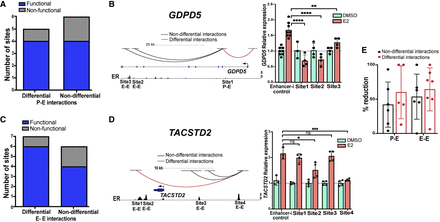
ERBSs in differential chromatin loops are subtly more important for the transcriptional response to estrogen. (A,C) The functionality of ERBSs based on Enhancer-i that are anchors of differential and nondifferential promoter–enhancer (P-E; A) or enhancer–enhancer (E-E; C) interactions is shown. (B,D, left) Genome browser tracks of GDPD5 (B) and TACSTD2 (D) show differential (red) and nondifferential (black) 3D genome interactions as well as ER ChIP-seq signal. (Right) Relative expression of GDPD5 (B) and TACSTD2 (D) normalized to DMSO when a candidate ERBS is targeted with Enhancer-i upon DMSO (green) or E2 (orange) treatment is displayed. Error bars represent SD. (****) P < 0.00001, (***) P < 0.001, (**) P < 0.01, and (*) P < 0.05, unpaired t-test; (ns) statistical insignificance. (E) The average percentage reduction is shown for each ERBS targeted with Enhancer-i split by the different types of 3D interactions. Each dot represents the average percentage reduction for one ERBS, and error bars represent SD.











