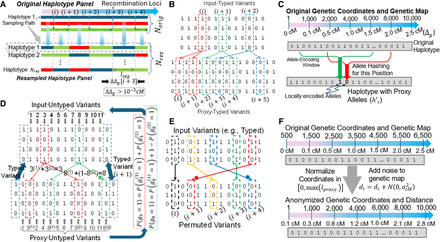
Illustration of proxy panel generation mechanisms. (A) Illustration of typed variant resampling. The original haplotypes are traced from left to right, and recombinations (indicated by switches between haplotypes) are randomly generated using the genetic map (depicted on
top). Recombinations are checked at the recombination loci shown with vertical dashed positions. The consecutive recombination loci have at least 0.001 cM genetic distance between
each other. ProxyTyper constrains the maximal segment length (lseg) to minimize chances of copying of long haplotype segments. (B) Typed variant augmentation. Given three consecutive typed variants (shown in dashed rectangles), each typed variant is copied
to a random position in the vicinity of itself. Each augmented proxy-typed variant is assigned the same genotypes of the original
variant (by default). After augmentation, the number of typed variants increased from three to six, adding three new augmented
variants. (C) Illustration of allele encoding (hashing). Given two windows (depicted with red and green windows), the allele on the proxy
haplotype is calculated as a combination of the alleles on the original haplotype. The two windows have independent encoding
functions. The function takes the alleles and the genetic distance as parameters to calculate the hashed (encoded) alleles
on the proxy haplotype (shown at the bottom). The proxy haplotype is calculated by encoding all windows. (D) Protection of untyped variants by haplotype partitioning. Two variants at positions 3 and 8 are partitioned to four new
proxy variants denoted by 3(1), 3(2), 8(1), and 8(2). The probabilities of imputed alleles for these variants are recovered from proxy variants. Denote the inversion of allele
on 8(2) (Methods) (E) Protection of untyped variants using (rolling) permutation between typed variant blocks. Untyped variant positions are randomly
shuffled between two consecutive typed variants. (F) Protection of the variant coordinates and the genetic distance. The coordinates are normalized to a preselected value lproxy, which obfuscates the typed and untyped variant positions. Genetic map is obfuscated using addition of Gaussian noise with
predefined variants 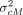 .
.











