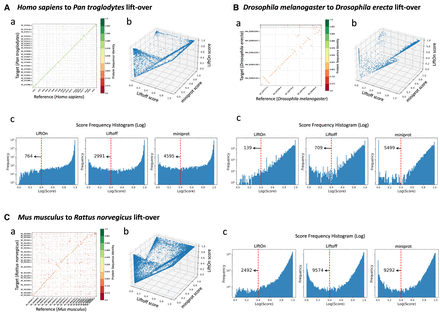
Evaluation of gene annotation lifting across species and tools using gene order plots, protein identity dot plots, and frequency distributions. (A) Comparative analysis of lifting over RefSeq v220 annotations from Homo sapiens (GRCh38) to Pan troglodytes (NHGRI_mPanTro3-v1.1). (B) Comparative analysis of lifting over annotations from Drosophila melanogaster (genome assembly release 6 + ISO1MT) to Drosophila erecta (Prin_Dsim_3.1). (C) Comparative analysis of lifting over annotations from Mus musculus (GRCm39) to Rattus norvegicus (mRatBN7.2). Graphs labeled a show protein-gene order plots, with the x-axis representing the reference genome and the y-axis representing the target genome. The protein sequence identities are color-coded on a logarithmic scale, ranging from green (one) to red (zero), and represent the degree of amino acid similarity, with one indicating identical sequences and zero indicating no shared amino acids. The gene order plot script was customized from LiftoffTools. Graphs labeled b are 3D protein sequence identity plots comparing Liftoff on the x-axis, miniprot on the y-axis, and LiftOn on the z-axis. Each dot represents a protein-coding transcript. If a dot is above the x = y plane, LiftOn's mapping produced a higher protein sequence identity score than the other programs. Graphs labeled c are frequency plots on a logarithmic scale of protein sequence identity for LiftOn (left), Liftoff (middle), and miniprot (right).











