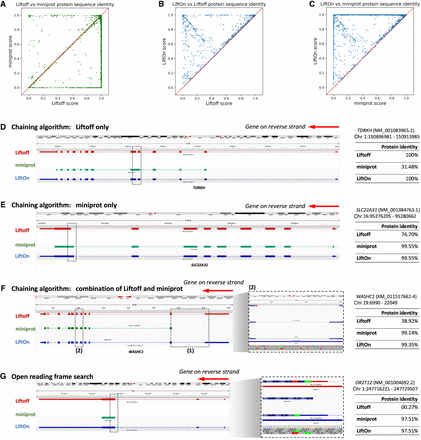
Comparative analysis of various tools for mapping RefSeq protein annotations from GRCh38 to T2T-CHM13 v2.0. (A–C) Scatter plots of protein sequence identity. (A) Comparison between miniprot (y-axis) and Liftoff (x-axis), (B) comparison between LiftOn (y-axis) and Liftoff (x-axis), and (C) comparison between LiftOn (y-axis) and miniprot (x-axis). (D–G) Examples of improved annotation owing to LiftOn's two-step PM algorithm. (D) LiftOn uses Liftoff's annotation to correct a splice junction missed by miniprot in transcript NM_001083965.2 of the TDRKH gene. (E) For transcript NM_001384763.1 of the SLC22A31 gene, LiftOn uses miniprot's annotation to resolve an incorrect acceptor site from Liftoff's annotation. (F) For transcript XM_011517662.4 of the WASHC1 gene, LiftOn combines both annotations to rectify an omitted CDS by miniprot and a misidentified splice junction by Liftoff between the fifth and sixth exons. (G) LiftOn's ORF search algorithm selects an alternative downstream start codon for a frameshift mutation in transcript NM_001004692.2 of the OR2T12 gene, thereby conserving the majority of the protein sequence.











