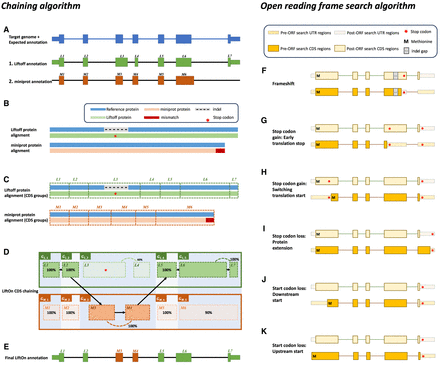
The protein-maximization (PM) algorithm consists of two modules: (A–E) the chaining algorithm and (F–K) the open reading frame (ORF) search algorithm. (A) Matched protein-coding transcripts mapped by Liftoff (green) and miniprot (orange) at the same location in a target genome.
The transcript in blue represents the correct transcript annotation on the target genome. Liftoff's mapping has an erroneous
splice junction between L3 and L4, whereas miniprot's mapping has a missing splice junction in M6. (B) Pairwise alignment results of the proteins mapped by Liftoff and miniprot to the reference protein. The figure shows a premature
stop codon in the Liftoff protein, and the miniprot alignment has a mismatched protein sequence at the end. (C) Pairwise alignment mappings with added exon/CDS boundaries. (D) CDSs are grouped based on the cumulative lengths of the amino acids in the reference protein as described in the main text.
In this example, CDSs are organized into groups: 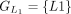 ,
, 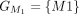 ,
, 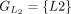 ,
, 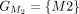 ,
, 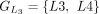 ,
, 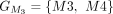 ,
, 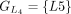 ,
, 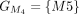 ,
, 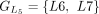 , and
, and 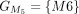 . The chaining algorithm iterates through each group, comparing the corresponding partial protein sequences to the reference
protein and chaining those with higher protein sequence identity. (E) In this example,
. The chaining algorithm iterates through each group, comparing the corresponding partial protein sequences to the reference
protein and chaining those with higher protein sequence identity. (E) In this example, 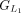 ,
, 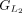 ,
, 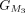 ,
, 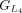 , and
, and 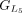 are chained, forming the new protein-coding transcript CDS list. This list includes L1, L2, M3, M4, L5, L6, and L7 in the LiftOn annotation. (F–K) Schematic diagrams illustrating how the ORF search algorithm handles various types of sequence mutations. This process leads
to changes in the gene annotation of both translated and untranslated regions (UTRs). (F) A frameshift mutation is a variation caused by the insertion or deletion of a sequence of nucleotides whose length is not
divisible by three. In this example, the indel introduces a premature stop codon. (G,H) Point mutations leading to premature stop codons. LiftOn searches for the longest ORF, considering two scenarios: G depicts the selection of the first encountered stop codon, and H illustrates the switch to the downstream start codon. (I) Stop codon loss. When a stop codon is deleted, LiftOn identifies a new stop codon in the 3′ UTR. (J,K) Start codon loss. In this scenario, LiftOn searches for a new start codon, exploring both downstream in the coding region
(J) and upstream in the 5′ UTR (K).
are chained, forming the new protein-coding transcript CDS list. This list includes L1, L2, M3, M4, L5, L6, and L7 in the LiftOn annotation. (F–K) Schematic diagrams illustrating how the ORF search algorithm handles various types of sequence mutations. This process leads
to changes in the gene annotation of both translated and untranslated regions (UTRs). (F) A frameshift mutation is a variation caused by the insertion or deletion of a sequence of nucleotides whose length is not
divisible by three. In this example, the indel introduces a premature stop codon. (G,H) Point mutations leading to premature stop codons. LiftOn searches for the longest ORF, considering two scenarios: G depicts the selection of the first encountered stop codon, and H illustrates the switch to the downstream start codon. (I) Stop codon loss. When a stop codon is deleted, LiftOn identifies a new stop codon in the 3′ UTR. (J,K) Start codon loss. In this scenario, LiftOn searches for a new start codon, exploring both downstream in the coding region
(J) and upstream in the 5′ UTR (K).











