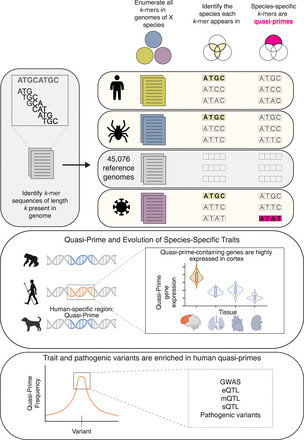
Identification of nucleic quasi-prime sequences in individual species. Schematic displaying the identification of a four-mer quasi-prime sequence in a species. All nucleic k-mers of a specific length are identified for each species, across all the reference genomes. k-mers that are shared between multiple species are removed, and only k-mers appearing in a single reference genome are kept, constituting the set of quasi-primes for that species for that particular length. As an example, we show a short toy sequence that is found in the human reference genome but is absent from all other species in our database, therefore constituting a human nucleic quasi-prime k-mer. Quasi-primes are associated with the evolution of human specific traits. For humans, quasi-prime-containing genes are enriched in the cortex and are associated with brain development and diseases. Human traits and pathogenic variants are significantly enriched in human quasi-prime sequences. Variants including expression quantitative trait loci (eQTLs), methylation QTL (mQTLs), splicing QTL (sQTLs), genome-wide association studies (GWAS) variants, and disease variants are more likely to be found in human quasi-prime sites.











