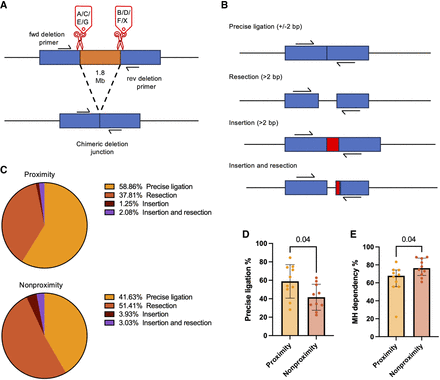
Five chimeric deletion junctions produced from each of the sgRNA pairs AB, CD, EF, and GX analyzed with Illumina MiSeq amplicon sequencing. (A) Definition of a chimeric deletion junction. (B) Characterization of sequencing reads. (C) Read distributions for the proximity and nonproximity sgRNA pairs (n = 10 sgRNA pairs in each group; six chromosome loci with one to two sgRNA pairs). (D) Precise ligation of chimeric deletion junctions for proximity and nonproximity sgRNA pairs (n = 10 sgRNA pairs in each group; six chromosome loci with one to two sgRNA pairs); P-value from Mann–Whitney U test. (E) Microhomology usage normalized to all reads with resection for proximity and nonproximity sgRNA pairs (n = 10 sgRNA pairs in each group; six chromosome loci with one to two sgRNA pairs); P-value from Mann–Whitney U test. The observations are biological replicates, and the error bars show the median and IQR.











