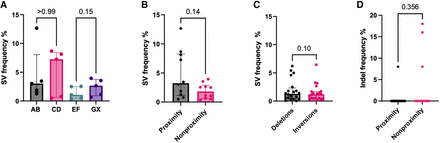
The effect of spatial proximity on SV frequency in H9 hESCs at the Chr 2, Chr 6, Chr 17, Chr 21, and Chr X loci. (A) SV frequencies for AB, CD, EF, and GX sgRNA pairs (n = 5 sgRNA pairs in each group; five chromosome loci with one sgRNA pair); P-values from Mann–Whitney U test. (B) SV frequencies between proximity and nonproximity sgRNA pairs (n = 10 sgRNA pairs in each group; five chromosome loci with two sgRNA pairs); P-value from Mann–Whitney U test. (C) Overall frequencies of deletions and inversions (n = 20 sgRNA pairs in each group; five chromosome loci with four sgRNA pairs); P-value from Wilcoxon matched-pair signed-rank test. (D) Individual sgRNA efficiencies by indel frequency (n = 20 sgRNAs in each group; five chromosome loci with four sgRNAs); P-value from Mann–Whitney U test. The observations are biological replicates, and the error bars show the median and IQR.











