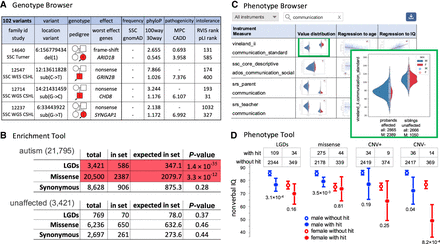
Query and statistical tools at GPF. (A) The genotype browser enables users to construct complex queries that involve the properties of genetic variants, the properties of affected genes, and the phenotypic properties of carrier individuals (Supplemental Fig. 1). Here, we partially present the results of a query for de novo LGD variants that fall within the FMRP target gene set and occur in children diagnosed with autism (for a description of the columns, see Fig. 1D). (B) The enrichment tool (Supplemental Fig. 2) enables users to test for enrichment of de novo variants within a gene set. The enrichment tool results for de novo mutations are shown in children with autism and unaffected children from the autism-related subset of the sequencing de novo “SD autism” in the FMRP target gene set. The top table shows that among 21,795 individuals with autism, 3421 de novo LGD mutations are found (N). Of these, 586 fall in FMRP target genes (O), although we expect to see only 347 (E). This corresponds to an enrichment with a significant P-value (pV), indicated with a red background. Missense mutations are similarly enriched in FMRP target genes. However, synonymous mutations are not enriched (white background). The bottom table shows the same analysis for unaffected individuals, which finds no enrichment for LGD, missense, or synonymous mutations. (C) The phenotype browser allows users to explore the available phenotypic data associated with a data set and download the subsets of interest. The panel shows a part of the search results for phenotypic measures related to “communication” in the SSC data set. The results are presented in a table, with one row per measure. For each measure, the table includes histograms of its values, separately in groups of individuals by gender, role, and affected status. The zoomed-in view (a feature provided by GPF) of the histograms plot for the communications_standard measure from vineland_ii shows the number of individuals for which the measure is available for the affected probands and the unaffected siblings separately by males and females and the histograms of the four groups (male and female probands and male and female siblings). The histograms show that the affected probands have diminished vineland_ii communication scores. The result table also displays the scatter plots and regression lines for each of the four measures against the individuals’ ages at assessment and their IQs. (D) The phenotype tool enables users to test whether a given phenotypic measure (e.g., nonverbal IQ) differs between SSC children who carry a specified type of genetic variant (e.g., de novo LGDs) and those who do not have such variants (Supplemental Fig. 3). The panel illustrates the impact of four de novo variant types (LGD; missense; CNV+, or large duplications; and CNV−, or large deletions) in genes with a low pLI rank (less than 1000) on nonverbal IQ. For each de novo variant type, the mean and 95% confidence interval are plotted for four groups of individuals: males and females with and without a de novo variant in the selected genes. There is a significant decrease in nonverbal IQ for affected children with de novo LGDs or CNV− within the genes with a low pLI rank. There is no effect of CNV+ mutations, and only a marginal effect is observed for missense variants. We note that panels B and D are stylized versions of content from GPF-SFARI, with reduced white space for better clarity. Supplemental Figures 2 and 3, respectively, illustrate the result representation generated by GPF.











