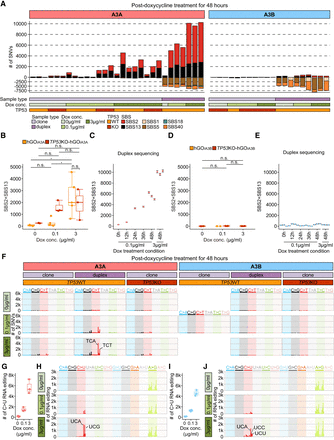
Mutational impact of APOBEC-associated single base substitutions in DNA and RNA. (A) Mutational burden of SNVs following APOBEC overexpression (A3A or A3B), measured by whole-genome sequencing of the clones and duplex DNA sequencing. The number of SNVs measured by the duplex DNA sequencing was normalized per diploid genome; (left) hGOiA3A lines, and (right) hGOiA3B lines. (B) Mutational burden of APOBEC-associated SNVs in the hGOiA3A and TP53KO-hGOiA3A clone sequencing. The number of A3A-associated SNVs (SBS2+SBS13) in hGOiA3A and TP53KO-hGOiA3A clones under each condition. Statistical significance was determined using a t-test: (*) P < 0.05, (n.s.) not significant. (C) Number of A3A-associated SNVs (SBS2+SBS13; normalized per diploid genome) in BotSeqS results for hGOiA3A lines under each doxycycline treatment condition. Black lines represent 95% confidence intervals based on a Poisson distribution. (D) Mutational burden of APOBEC-associated SNVs in the hGOiA3B and TP53KO-hGOiA3B clone sequencing; the number of A3B-associated SNVs (SBS2+SBS13) in hGOiA3B and TP53KO-hGOiA3B clones under each condition. Statistical significance was determined using a t-test. (E) Number of A3B-associated SNVs (SBS2+SBS13; normalized per diploid genome) in BotSeqS results for hGOiA3B lines under each doxycycline treatment condition. Black lines represent 95% confidence intervals based on a Poisson distribution. (F) Mutational burden and spectra of APOBEC-associated SNVs in each experimental condition. The number of SNVs in BotSeqS results were normalized per diploid genome; (left) hGOiA3A lines, and (right) hGOiA3B lines. (G) Number of C > U RNA editing in bulk RNA-seq in hGOiA3A lines (n = 3 per condition), normalized per 3.1 Gb of mapped bases. (H) Spectra of RNA editing in trinucleotide contexts in hGOiA3A lines. (I) Number of C > U RNA editing in bulk RNA-seq in hGOiA3A lines (n = 3 per condition), normalized per 3.1 Gb of mapped bases. (J) Spectra of RNA editing in trinucleotide contexts in hGOiA3B lines.











